Hybrid Sequencing and Map Finding (Hysemafi) Optional Strategies Review
Abstract
Using second-generation sequencing (SGS) RNA-Seq strategies, extensive alterative splicing prediction is impractical and high variability of isoforms expression quantification is inevitable in organisms without truthful reference dataset. nosotros report the evolution of a novel analysis method, termed hybrid sequencing and map finding (HySeMaFi) which combines the specific strengths of third-generation sequencing (TGS) (PacBio SMRT sequencing) and SGS (Illumina Hi-Seq/MiSeq sequencing) to effectively decipher gene splicing and to reliably estimate the isoforms abundance. Error-corrected long reads from TGS are capable of capturing full length transcripts or equally big partial transcript fragments. Both true and faux isoforms, from a particular gene, as well equally that containing all possible exons, could be generated by employing different associates methods in SGS. We beginning develop an effective method which can constitute the mapping relationship between the mistake-corrected long reads and the longest assembled contig in every corresponding cistron. According to the mapping data, the true splicing pattern of the genes was reliably detected, and quantification of the isoforms was as well effectively adamant. HySeMaFi is also the optimal strategy by which to decipher the full exon expression of a specific gene when the longest mapped contigs were called equally the reference prepare.
Introduction
Short-read second-generation sequencing (SGS) has get a powerful tool for the description of gene expression levels and individual splice junctions in those organisms with a reference genomeane,2,3. Despite the availability of verified and improved algorithms along with skilful software options, information technology is yet hard to identify full-length transcript isoforms using the SGS information. Thus, it is not yet possible to get a complete understanding of all spliced RNAs within a transcriptome using SGS, fifty-fifty in those organisms with a reference genome4. Given that, even with a true reference sequence, spliced RNAs can only be partly inferred from a patchwork of short fragments, information technology is non surprising that, to date, in that location are no reports of the successful performance of gene splicing analyses by SGS in organisms lacking a reference genome. At the same fourth dimension, it is well recognized that reconstruction and quantification of transcript isoforms from short-read sequencing is insufficiently accurate5,6.
Transcriptional and alternative splicing events may be detected, to some extent, in RNA-Seq data from SGS. However, total-length mRNA isoforms are not directly captured or may fifty-fifty non be detected, despite the employment of diverse powerful computational algorithms and software. Pacific Biosciences (PacBio) developed a novel platform enabling single molecule real fourth dimension (SMRT) sequencingseven, and this represented an advance over SGS, i.east. third-generation sequencing (TGS). TGS, although currently having the limitations of reduced raw accuracy and lower throughput, has significant capacity for awarding in de novo sequencing, and may also assistance the assay of linkage of culling splicing forms and of variants across long amplicons, as has been successfully used in the study of the human genome and of other organisms with an available reference genome4. In the by, several approaches take been developed that use short high-identity sequences to correct the error inherent in long unmarried-molecule sequences, and thereby generate a highly accurate hybrid consensus sequenceeight,9. With the development of improved correction of base of operations-call errors in TGS, the technique is increasingly widely used in genome sequencing due to its reward of generating long reads10,11,12. To date, the advantages of SMRT sequencing have been utilized in guild to identify extensive alternative splicing patterns and as well thousands of transcript isoforms from numerous organisms which have a known reference genome13. However, no strategy or methodology currently exists that tin extensively decipher gene splicing or efficiently calculate the expression of alternative isoforms on a large scale in an organism for which the genome sequence is not available.
Many eukaryotic genes exhibit extensive culling splicing, and alternative splicing (Equally) is of importance as information technology represents a posttranscriptional mechanism that can mediate regulation of cistron expression14,15. The high charge per unit of occurrence of AS in plants during unlike developmental stages or when growing under various environmental conditions has sparked a growing interest in AS as a regulatory mechanism, and has led to further studies focused on revealing the total extent of AS by deep sequencingsixteen,17. It is our understanding that, using the analysis techniques currently available, information technology is not possible to routinely and accurately decipher the Equally profile based solely on SGS if a detailed genome note is non bachelor. SGS is capable of generating a large number of short reads to support the databases produced by electric current RNA-Seq engineering science. Based on short reads, the following information tin exist obtained: (i) frequency of reads mapped to a contiguous genomic segment (exonic reads), and (2) frequency of reads mapped to 2 contiguous segments of the genome with a single gap (junction reads)18,19,20. Information technology is possible to infer isoform-specific expression from exonic reads and junction reads if the total set of possible isoforms for the given cistron is already established. However, such isoform quantification presents significant difficulties if the ready of isoforms is not known or only partially known due to insufficient lengths of the SGS reads21. Thus, taking mentions above, information technology is possible to deduce a methodology that could extensively decipher gene splicing and effectively compute gene expression profiles using close-to-real levels of isoforms.
In this paper, nosotros initially extracted iv singled-out tissue types in social club to generate extensive data sets using TGS, SGS and MiSeq methods. We used these datasets to develop a novel assay technique which is based on the ability of mistake-corrected long reads in TGS to capture many transcripts in full length or, at to the lowest degree, every bit big partial transcript fragments. Furthermore, the various alternative contigs, including the true and false forms and too that containing the total complement of exons of a item gene, can be generated by employing the unlike suitable associates methods in SGS. Based on these information outputs, we take adult an effective mapping method, coupled with an associated analysis pipeline, which we demonstrate is able to constitute the mapping relationship betwixt the fault-corrected long reads and the longest assembled contig in every corresponding gene. The mapping method was also tested and verified using the existing annotated isoform sets and the contigs datasets assembled from SGS past de novo assembly in Arabidopsis. Secondly, relying on the mapping technique, the longest contigs, typically comprising the full complement of exons respective to individual genes, were selected to form a reference library, and and so from this, all-encompassing gene splicing patterns were adamant. The alternative splicing sites were verified by datasets from MiSeq. Thirdly, past utilizing the corrected long reads of isoforms in TGS or the mapped longest contigs from SGS as reference materials, and combining the alignments with a large number of short reads from unlike samples, the expression quantification of isoforms of genes was also finer determined. In summary, a novel optional strategy, coupled with its analysis pipeline, were developed and demonstrated, which allows efficient deciphering of gene splicing and isoform expression in organisms whose genome is unavailable; this method is described as hybrid sequencing and map finding (HySeMaFi).
Results
Principles of hybrid sequencing and map finding (HySeMaFi)
The basic principle of the HySeMaFi technique is outlined in Fig. 1 (and detailed in Supplementary Fig. S1). The method centers on the SGS and TGS technologies and the mapping between the single-molecule corrected PacBio reads and the assembled contigs derived from SGS. It is hypothesised that if the A and B isoforms from a given gene truly exist in specific prison cell, tissue or organ types, and then the application of two sequencing strategies volition generate ii singled-out transcript datasets (Fig. 1). Through the SGS and associates technique, it is theoretically unavoidable that diverse different molecule sets, containing the true molecules and/or false ones, could be generated when employing different assembly ciphering methods with alternative parameters, such as using shorter K-mer and low coverage values (Fig. i). Thus the longest molecule (i.e. including all exons of the gene) was assembled by combining all the other real, besides as false, molecules that were generated in a specific tissue type (Fig. 1 and Supplementary Fig. S2). If all genes were considered, then more alternative molecules were obtained and there was a loftier level of confidence that the assembled contig pool contained the longest molecule of every cistron, as well as the other real isoforms of each (Fig. 1). In TGS, PacBio reads, afterward correction for brusque reads or self-alignments, represent full-length or most full-length true transcripts (Fig. ane and Supplementary Fig. S2). Therefore, it is entirely feasible that nosotros could constitute the mapping relationship between the longest molecule in SGS and the corrected PacBio reads (Fig. 1 and Supplementary Fig. S2) using suitable alignment methods. Thus, taking the corrected PacBio long reads and the longest contig into consideration, this scheme shows great potential to extensively decipher gene splicing by reasonable alignment analysis (Fig. 1 and Supplementary Fig. S2). In improver, it is also apparent that, if the longest contigs from SGS or the corrected PacBio long reads from TGS are employed every bit reference input data, the expression abundance of the factor or of each isoform, specific to a given tissue, will exist effectively calculated (Supplementary Fig. S1). Thus, patterns of gene splicing and expression may be effectively determined through this approach, even in those organisms without an available reference genome.
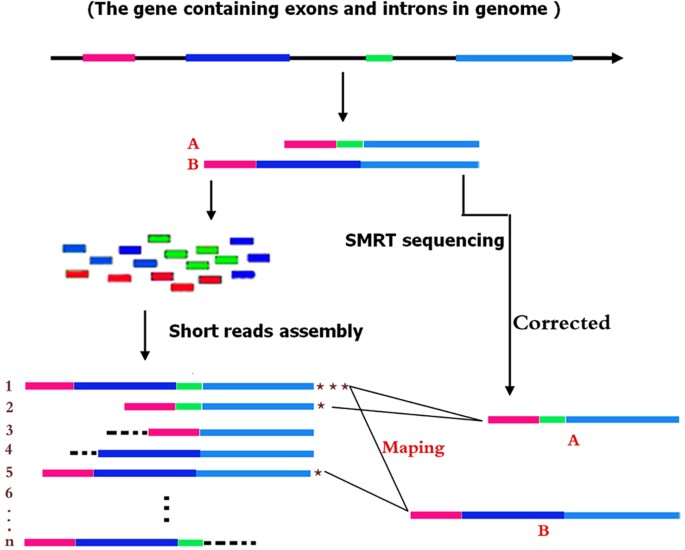
Based on each factor containing exons and introns, it is hypothesised that (A,B) isoforms are generated in particular tissues, and and so 2 sequencing strategies are employed to yield the multiple molecules. In SGS, it is may be that all possible molecule forms are produced when employing unlike assembly methods with various parameters, wherein the real (true) molecule is contained. In comparison, in single molecule real-fourth dimension (SMRT) sequencing (TGS), only two tissue-specific real molecules (after correction) were generated. It is possible to map the molecules between SGS and TGS molecular forms using our developed mapping methods. Finally, the molecule containing all of the gene exons in SGS is detected and used to follow downstream analysis.
Contigs derived from SGS
The sequencing of 12 Petunia hybrida cDNA libraries, derived from roots, stems, leaves and flowers, generated a full of 945,501,540 clean reads (Accession number SRR4116645–56 at NCBI) after removal of adapter sequences and low quality reads, and each of the libraries yielded high quality reads in the range 63–84 M (Supplementary Tabular array S1). To maximize transcript coverage, we pooled all of the clean Illumina reads together in order to perform de novo transcriptome assembly using the Trinity assembler employing various parameters. To obtain the maximum number of assembled theoretic nucleotide molecules, we employed either low threshold or default parameters in the assembly process and consequently produced two dissimilar de novo transcriptome versions containing 490,981 and 412,941 transcripts (genes), respectively (Fig. 2a and Supplementary Table S2). The statistics describing these assembled data sets are given in Tabular array S2. The average contig length that was assembled using Trinity under depression threshold parameters was 1648.xv bp and the N50 value was 2930 bp; this compares with the respective values of 1394.38 bp and 2579 bp when default parameters were used in the assembly procedure (Fig. 2b and Supplementary Table S2). Comparative assay indicated that longer molecules (≥3000 bp) were produced when the custom parameters were employed (Fig. 2c). In MiSeq, more than x Chiliad high quality paired-end MiSeq reads were accomplished (Supplementary Table S3). After processing, we obtained iv,596,458 extended fragments with an boilerplate length of 444 bp (Supplementary Table S4), and the distribution of extended fragments length ranged between 300–590 bp (Supplementary Fig. S3). In additional, clean reads after removal low quality reads in Arabidopsis were summarized as Table S5, and ii different de novo transcriptome versions generate 44,934 and 44,914 genes (Supplementary Table S6).
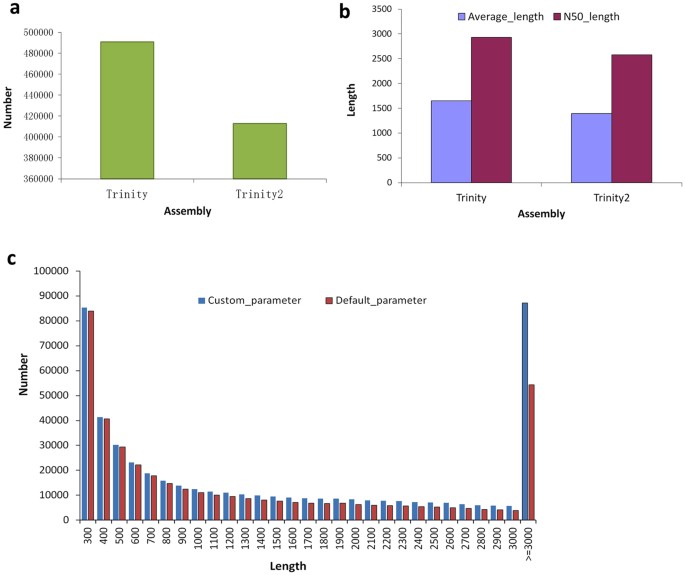
(a) The number of assembled contigs by Trinity assembler using low threshold or default parameters. (b) Average and N50 length of contigs under the two different assembly parameters. (c) Distribution of the number of contigs at various lengths.
Long, corrected PacBio reads derived from TGS
Based on the two SMRT sequence libraries, six SMRT cells were employed to generate the PacBio raw sequence data. The number of insert reads in each SMRT cell ranged from 46k to 54k (Supplementary Table S7), with an average of >ninety% quality. In total, later on the nomenclature assay, nosotros obtained 299,542 reads of inserts of which approximately 53.7% (i.e. 160,728) represented full-length reads (Accession number SRR4117145-46 at NCBI) after raw data processing (Supplementary Table S8). Moreover, the number distribution for insert length across the various reads indicates that more than 20 Thou insert-reads were obtained (Supplementary Fig. S4a) and nearly of these were of a high quality (Supplementary Fig. S4b). For the full-length non-chimeric reads, the distribution of read lengths was consequent with the level of transcript lengths seen in other establish species (Supplementary Fig. S5). In our study, nosotros adapted three strategies in order to reach the high quality PacBio-read dataset: namely, (1) full-length reads with an accurateness greater than 99% were isolated after removing reads shorter than 200 bp; (2) consensus isoforms were predicted using Ice software so polished using Quiver software, after the removal of any reads shorter than 200 bp or with a predicted accurateness lower than 75%; (iii) the total length transcripts were corrected by using LSC with SGS brusk reads. In the LSC correction process, about of the full length transcripts were supported by the SGS short reads, and a coverage value of more than 99% represented the highest number of short reads (Fig. 3a). Afterwards correction, very few differences were seen between the corrected PacBio reads derived from consensus information and the chief PacBio reads (Fig. 3b). At the end of the analysis process, nosotros obtained a total of 160,293 mistake-corrected long reads. Indistinguishable long reads were removed by undertaking the clustering based on map finding according to our algorithm, and a total of 85,571 unique long reads were generated (Fig. 3c). A comparison to the transcripts assembled by SGS showed that the distribution of lengths of the corrected PacBio long reads appeared to more typically resemble the true transcript lengths (i.e. 1–3 Kb) seen in plants. This is significantly different to the transcriptome data assembled from SGS with regard to the frequency of long transcripts (Fig. 3d).
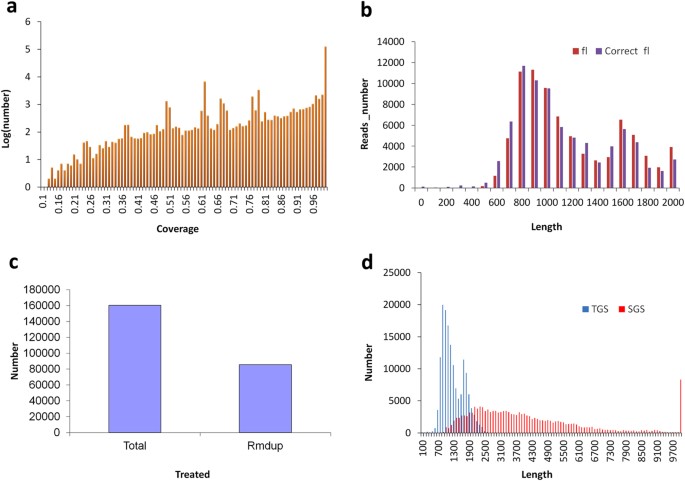
(a) PacBio corrected long reads supported by the Hiseq information with varied coverage. (b) Numbers distribution of PacBio long reads before and after correction using LSC. (c) Number of PacBio corrected long reads before and afterwards duplication-removal using our adult methods. (d) The number distribution of PacBio corrected long reads and Hiseq assembled contigs at various lengths.
Contigs Mapping between corrected PacBio reads and contigs from SGS
Using the LSC corrected PacBio reads as the query sequences, the contigs from SGS (Illumina Hullo-Seq) were aligned by Trinity using default parameters, and a full of 85,571 unique reads were mapped to specific transcripts according to our novel redundancy-removal and mapping method. Our process does not rely on generalized identifications based on factor structures and, therefore, is relatively unbiased. Of these mapped PacBio reads, fifty% (43,132) had a more than 99% identity charge per unit (Fig. 4a). To test our novel analysis method, a further two corrected data sets derived from TGS were employed to perform the mapping analysis. Using the consensus isoforms predicted using ICE software and then polished by Quiver every bit the query sequences, it was observed that over 13 chiliad, or 12 yard of the PacBio long reads could exist mapped to the transcripts assembled using Trinity when employing default, or low stringency parameters, respectively, at the more than 99% identify threshold (Fig. 4b). However, using the remaining total-length reads with an accuracy level greater than 99% every bit divers by TGS sequencing, few of the long reads could be mapped to the Trinity-assembled transcripts (Fig. 4c). It is clear that the higher error charge per unit in SMRT sequencing persists despite the removal of low quality data. In comparison, of the approx. 491,000 transcripts assembled in SGS, more 33% of transcripts could not be mapped to the PacBio corrected long reads, and simply 55,000 transcripts (11.thirty%) could exist mapped to the PacBio corrected long reads with a 99% identity level (Fig. 4d). This indicates that a big number of untrue transcripts are unavoidably generated as part of the SGS assembly process. In Arabidopsis, total existing 19194 annotated isoforms could map to specific transcripts from SGS by de novo assembly just using 3 downloaded datasets according to our novel back-up-removal and mapping method, of which full 13336 transcripts contains full exons (Fig. 4e). In those non-mapping genes, information technology exists many differences in exon number (Fig. 4f).
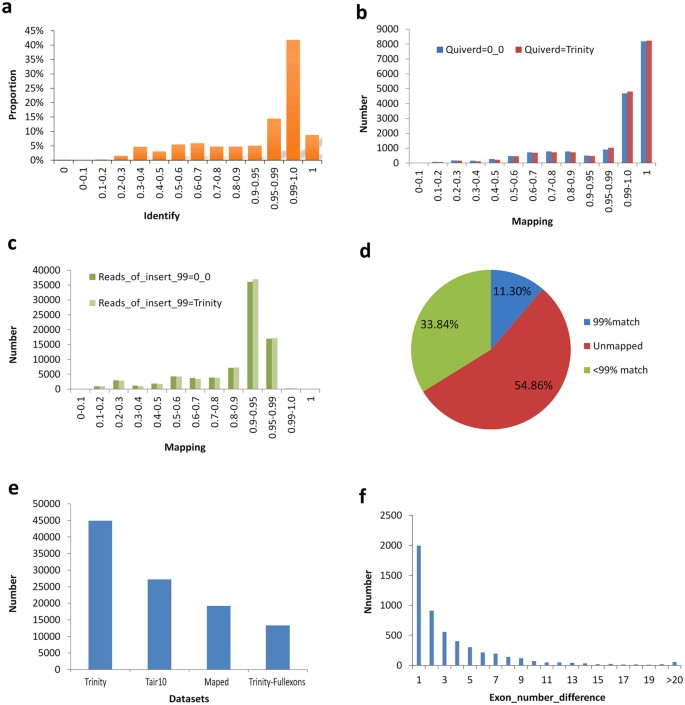
(a) The mapping between PacBio corrected long reads using LSC method and the assembled contigs using low threshold parameters at diverse identity rates. (b) The mapping between PacBio corrected long reads using ICE and Quiver software, and the assembled contigs using depression threshold or default parameters. (c) The mapping between PacBio corrected long reads with a more than 99% corrected rate, and the assembled contigs using low threshold or default parameters. (d) Pie chart of mapped or unmapped analysis to the contigs every bit assembled in total from SGS by Trinity software with a 99% level mapping threshold. (eastward) The mapping between the existing annotated isoform sets and the contigs datasets assembled from SGS by de novo assembly in Arabidopsis. (f) The exon number divergence between the existing annotated isoform sets and the contigs datasets assembled from SGS in Arabidopsis in many mapping.
Gene culling splicing detection
To effectively identify the alternatively spliced isoforms of genes, and confirm that they could be verified past different lines of testify, we first performed PacBio long reads clustering analysis. By and large, when gene alleles and associated homologs were grouped against these results they typically shared the same alternative splicing patterns. The results of our clustering analysis showed that more than than fourscore% of isoforms were grouped according to two types of molecule, merely there were as well more than than 100 clusters that contained over 50 molecules (Fig. 5a). This result shows that varied isoforms, generated by a single factor, were widely found in our test samples. Based on our mapping methods and using the longest transcripts as reference data, nosotros found that, in addition to the gene isoforms corresponding to the full complement of possible exons, at to the lowest degree ii,264 genes showed more than two alternative splice forms (isoforms). The bulk of these genes corresponded to ii-to-three isoforms, and this scenario is judged to exist a reasonable representation of the truthful alternative splicing pattern in plants (Fig. 5b). Furthermore, nosotros identified 498 genes that displayed at least three culling splicing patterns (Supplementary Table S9), and in the majority of these cases the homologs of the genes take been previously reported to have alternative splicing patterns. In a parallel assay, nosotros used the Miseq information as the query fix against which to marshal the longest and mapped contigs assembled in SGS, and the mapping results showed a high degree of consistency to the long reads data sets for the majority of genes analyzed (Fig. 5c and d). In improver, gene alleles sharing the same splicing pattern were also detected when the alignment was performed using BLAT with the similarity or identity level set below 100% (Fig. 5c,d).
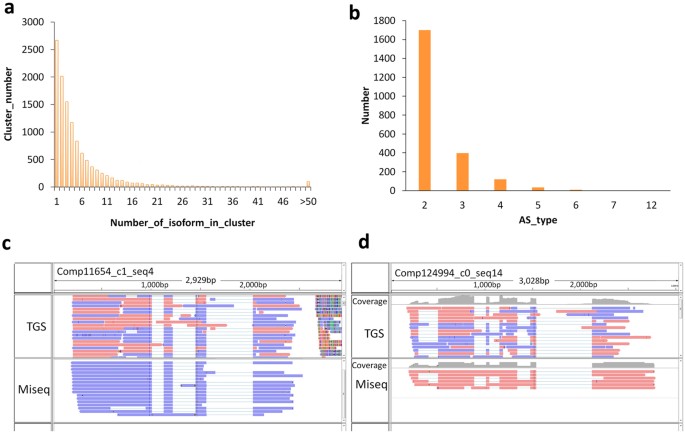
(a) Clustering of the SGS assembled contigs (genes) mapped past the PacBio corrected long reads. (b) The statistics of the gene numbers with various alternative splicing forms. (c,d) Cases selected to verify the effectiveness of detecting the isoforms (alternatively spliced molecules) using the HySeMaFi method, equally are supported by the Miseq data (run across arrow).
Gene expression defined past second sequencing using PacBio contigs as reference
To take account of the different expression levels of root-specific isoforms, the total number of 85,571 LSC corrected PacBio reads was used as the reference dataset. Information technology was shown that 2,904, one,618 and 3,868 private isoforms had significantly higher expression levels in roots as compared to those in flower, stem and leaf tissues, respectively; of these, 639 transcript forms were consistently expressed well-nigh highly in roots (Fig. 6a). A heat map illustrates the expression of these 639 genes which was significantly (at least twofold) and consistently higher across triplicate root samples, as compared to triplicate samples of other tissues (Fig. 6b). On the other mitt, 1,967, 1,219 and 2,780 isoforms had significantly lower expression levels in roots equally compared to those in flower, stem and foliage tissues, respectively. Of these, 869 were consistently expressed at the everyman levels in roots, and this expression pattern was robustly supported for all of these genes by heat map assay (Fig. 6c,d). In the traditional RNA-seq analysis which was based on the 490,981 total number of transcripts assembled in SGA, after the standard clustering analysis and removal of redundant sequences, 193,749 transcripts were finally used to make the reference dataset. Co-ordinate to this analysis, 896 transcripts were specifically expressed more highly in roots (Supplementary Fig. S6a) only a rut map shows that the significantly college expression levels of these genes were not seen consistently across three unlike root samples (Supplementary Fig. S6b). In improver, 666 transcripts were found that were specifically expressed at lower levels in roots (Supplementary Fig. S6c,d). Comparing of the ii analysis methods demonstrated that highly reproducible results, whether concerning upward- or down-regulation, were obtained using our novel calculation methods and with corrected PacBio reads supplying the reference sequences. When comparing the mapping relationship between PacBio corrected long reads and contigs assembled from SGS, it should be considered that some transcripts would have been removed during the clustering steps, so the expression of mapped contigs may not correlate (Fig. 6e,f). If the same molecule is present in the PacBio corrected long reads and also in the contigs assembled from SGS, information technology should be presented with the same expression pattern in each analysis. Truthful (Fig. 6g) or false molecules (Fig. 6h) in SGS were employed in the reference dataset in standard RNA-Seq assay. Furthermore, abundance estimates for most of the highly-expressed isoforms from individual genes were entirely accurately predicted when taking the corrected PacBio reads equally the reference dataset (Supplementary Fig. S6e,f).
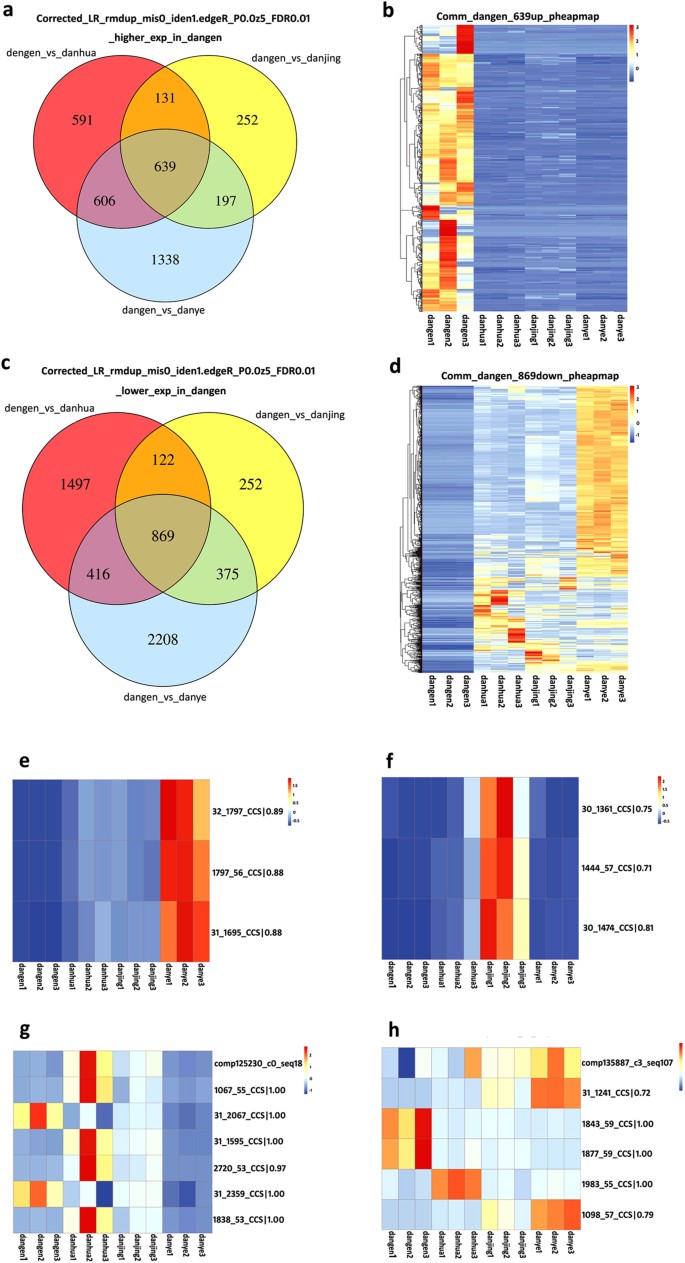
(a) The higher expression of isoforms specifically in roots; (b) Rut map shows the expression of 639 genes; (c) The lower expression of genes specifically in roots; (d) Heat map shows the expression of 869 genes; (e,f) The existing isoforms were detected to be differentially expressed in the tested samples, and the mapped contigs had been removed in the clustering analysis in the traditional RNA-Seq assay; (k,h) The existing isoforms were detected to be differentially expressed in the tested samples and the mapped contigs had as well been institute to exist differentially expressed.
Discussion
Hybrid sequencing and map finding involves a novel strategy to decipher gene splicing and expression
The hybrid sequencing and map finding strategy (HySeMaFi) described hither is based on the following technical points. (1) SGS is employed and the assembly is conducted using depression stringency settings for parameters such as Ker and coverage value. (two) SMRT sequencing is employed and the reads are corrected with short reads or other corrective methods. (three) A suitable method is used to plant the mapping relationship betwixt assembled contigs and the PacBio corrected reads, and the longest molecules i.east. those containing all exons of a given factor are identified. (4) Using these longest molecules and PacBio corrected reads as input data, it is possible to determine culling splicing patterns of genes past employing suitable alignment methods. (v) Using PacBio corrected reads as reference sequences confronting which to perform the RNA-seq analysis, it is possible to effectively illustrate the dissimilar expression patterns of various isoforms.
With regards to the traditional RNA-seq analysis, it should be noted that in order to get reliable information of a factor'southward alternative splicing patterns, a contiguous genomic segment should be mapped, and there remains substantial difficulties with isoform identification and quantification19,20,21. Here, using the hybrid sequencing and map finding method nosotros were able to obtain the longest contigs (i.eastward. those containing all exons/corresponding to the full-length gene without introns) by a similarity assembly approach to genome assembly. Therefore, using this method it is possible to perform corrected gene alternative splicing analysis, isoform identification and quantification analysis, and this arroyo may even facilitate greater accuracy in other downstream analyses. Our strategy takes reward of the library constructions from outputs of brusk-read adjacent-generation sequencing and achieves quantification of expression by calculating the mapped frequency of reads, whilst in addition taking advantage of the molecular abyss of the data from single molecule real-time (SMRT) sequencing (Fig. 1). Thus, the hybrid sequencing and map finding method involves a novel strategy to overcome the difficulties of de novo isoform discovery and can also remedy the insufficiencies of curt-read sequencing with regard to accuracy of construction and quantification of isoforms5,6.
Extensive mapping exists betwixt the corrected PacBio read and contigs assembled from short reads
Theoretically, it is clear that various molecules, including both real and false forms, can be generated past employing dissimilar assembly methods based on short reads. Our study was able to verify this premise (Fig. 2) with the identification of the longest molecule for each gene, consisting of all exons, being assembled in combination with all other real as well equally imitation molecules (Supplementary Fig. S2). In many cases, the longest of the short-reads assembled molecules is quite close to the full-length cistron and has the highest similarity to the factor when compared to all of the isoforms derived from single molecule real-time sequencing. Thus, the longest molecules assembled from short reads take the potential to be used as reference sequences in subsequent analysis pathways when the reference genome is not available, every bit is the example for the majority of organisms. Through the apply of suitable alignment methods, it is completely feasible that the longest molecules (i.e. those containing all of a gene'southward exons), as assembled past curt reads, could be used to map all of the real molecules (from the corrected PacBio reads) which contain all or some of the gene exons. Theoretically, the dataset of the longest molecules can play the role of a reference genome in mapping and too other subsequent assay pathways (Fig. 1 and Supplementary Fig. S2). At that place are many available alignment methods, of which BLAT is commonly used to detect regions in a corresponding genomic sequence which are similar to the query sequence and thus, determine the distribution of exonic and intronic regions of a cistron. In our study, using the BLAT method and applying a high identity score, it was shown that extensive mapping existed between the corrected PacBio reads and the contigs assembled from short reads, although various corrective strategies were employed in the raw PacBio RS reads. In additional, the mapping method was tested and verified using the existing annotated isoform sets and the contigs datasets assembled from SGS in Arabidopsis.
Alternative splicing patterns of genes are finer detected past Hybrid sequencing and map finding
Alternative splicing is a mechanism by which multiple proteins can be produced from a single cistron, and it is also a posttranscriptional mechanism that can regulate cistron expression22. The corrected PacBio reads derived from single molecule real-fourth dimension (SMRT) sequencing have the advantage that they contain all of the information originating from a single RNA molecule, and thus this information can be sufficient to notice factor splice sites5,23. Based on the genome information, NGS brusk-read information can identify splice sites by SpliceMap or Tophat, withal, the largely incomplete and uncorrected assembled transcripts can substantially impede the direct identification of distinct isoforms. Thus, to appointment, there are no reports that certificate the successful use of NGS short-read data to identify splice sites in organisms as a mode to over-come a brusque-autumn in genome information. SMRT long-reads tin exist used as a way to detect isoforms effectively and the data has been employed to reveal the corresponding culling splicing events in many organisms with an bachelor reference genomeiv,21,24. To effectively decipher alternative splicing patterns in organisms lacking bachelor genome information, nosotros have developed the hybrid sequencing and map finding method and take demonstrated that information technology can directly identify distinct isoforms of individual genes (Fig. 5). Here, in accord with our initial hypothesis, the longest contigs assembled by curt reads were constitute to be about to, or the same as the complete cistron, providing the total isoform sequences covered the full genomic department corresponding to the gene (Supplementary Fig. S2). To verify the culling splicing patterns detected by the hybrid sequencing and map finding technique, we aligned the MicroSeq data, which presented the actual sequence information at 400–500 bp lengths, to the longest contigs assembled by sequencing brusque reads. We found that almost all of the splicing sites detected were supported by the MicroSeq sequencing data (Fig. 5). Furthermore, we used the longest contigs that were detected to accept at least four isoforms as the query sequences and conducted BLAT analysis of a non-redundant (NR) database. It was plant in the majority of cases that the homologous genes were already reported to present comparable alternative splicing patterns (Supplementary Table S2). Thus, it is a robust finding that the events, such equally exon skipping, intron retentivity and isoforms identification, are effectively detected past the hybrid sequencing and map finding method, peculiarly in those deepen SGS and TGS cases. Certainly, information technology too nowadays the limitation that the culling splice borders are not well distinguished since it is curt of those real intron information that is not transcribed.
Tissue specific isoforms of genes were effectively deciphered by using the corrected PacBio reads as reference
In the calculations of isoform-specific gene expression based on SGS data, a short read consequent with 2 or more isoform types is unremarkably regarded to be generated from the more than abundant isoforms25. Isoform affluence estimates based on SGS data are highly sensitive to correction calculations and the true number of bachelor isoform datasets21. In traditional RNA-Seq analysis, the two types of reference isoform are usually sourced from the existing libraries when the genome is available or, alternatively, from candidate isoform sets assembled by SGS analysis tools. RNA-seq assay conducted using the existing isoform libraries as reference tends to increment the variability of the abundance estimates of the expressed isoforms since not all isoforms in the existing reference libraries are truly expressed in the tested sample, or alternatively, the reference library perhaps incomplete. For RNA-seq using the isoform libraries assembled from brusque reads as reference, a candidate isoform prepare may include numerous simulated isoforms, so differing essentially from the true number of isoforms21. Thus, utilise of the fault-corrected long reads is optimal for selecting corrected and true isoform datasets every bit the reference for a sample, and information technology volition also enable much more than reliable isoform quantification from SGS reads. In our study, we performed SMART and SGS sequencing for tissues of the roots, stems, leaves and flowers of P.hybrida, and used office of the SGS data to correct the PacBio long reads. These corrected PacBio long reads were and so utilized as reference isoform libraries. In addition, we besides performed traditional RNA-seq analysis with no reference sequence background. Our findings clearly showed that either elevated or depressed expression patterns in roots were both effectively detected using the corrected PacBio long reads every bit the reference isoform library (Fig. 6a,b). Furthermore, for the detected isoforms with an elevated root-specific expression, the majority of the homologous genes in specific species are known to likewise show high or low expression in root tissues (Supplementary Tabular array S10 and Table S11). By dissimilarity, use of the traditional RNA-Seq analysis method revealed that the longest assembled contigs mapping to the PacBio corrected reads may, or may non, truly be present in the concluding reference isoform library post-obit clustering analysis of the assembled transcripts. Thus, it is inevitable that determining isoform expression levels using traditional RNA-Seq substantially increases the doubt of the abundance estimates of the expressed isoforms.
In summary, we have presented an optional strategy past which to extensively decipher gene splicing and expression by hybrid sequencing and map finding in organisms without a reference genome. In this method, mapping analysis is carried out between the corrected PacBio long reads and the contigs assembled from short reads (which correspond to all the isoforms of an private factor), and the longest contig assembled from the short reads data is selected to group within the longest contig dataset. This latter dataset is used every bit the reference against which to detect the culling splicing patterns of every factor past using reasonable alignment strategies. Additionally, however, by using redundancy-deducted PacBio long reads as established reference libraries, it will be possible to profoundly increase the accuracy of the abundance estimates of the expressed isoforms.
Materials and Methods
Plant materials and sample training
P. hybrida plants were grown outside in the experimental field of Huazhong Agricultural Academy, Wuhan, China. Roots, stems, leaves and flowers of 9 plants were evenly harvested respectively for diverse sequencing library constructions. All samples were frozen in liquid nitrogen and stored at −fourscore °C until required for analysis.
Hiseq and Miseq library structure and sequencing
Total RNA was isolated with TRIzol reagent (Invitrogen) and mRNA was purified using oligo (dT) magnetic beads (Dynabeads) according to the manufacturer'due south instructions. Approximately 100–200ng of PolyA RNA was fragmented to perform first-strand cDNA using reverse transcriptase and random hexamer-primers. The second-strand cDNA was synthesized using DNA polymerase I and RNaseH. The cDNA was end-repaired and A-tailed, and Illumina paired-cease adapters were added. Afterward size option on an agarose gel and PCR amplification, samples were sequenced on the Illumina Hi-Seq 2000 system, generating paired-end (PE) reads with a length of ii × 100 bp. Samples were sequenced on the Illumina Mi-Seq arrangement, generating paired-stop (PE) reads with a length of 2 × 300 bp.
PacBio library structure and sequencing
SMRTbell libraries were synthetic by using the compatible mixed RNA course roots, stems, leaves and flowers and Pacific Biosciences' 1.0 template prep kit (office 100-259-100) according to the manufacturer's instructions. The synthesized cDNA was run on an agarose gel and two separate size ranges were fractionated: 1–ii kb, 2–three kb. Each size fraction was extracted from the gel and treated according to Pacific Biosciences' template preparation and sequencing protocol. The Deoxyribonucleic acid/polymerase binding kit P5 (part 100-256-000) and v2 primers were used to make SMRTbell templates bound to polymerases. The polymerase–template complexes were bound to magbeads using Pacific Biosciences' Magbead binding kit (role 100-133-600), and SMRT sequencing was then carried out on Pacific Biosciences' real-time sequencer RTII by using DNA Sequencing Reagent 3.0 (part 100-254-800). All movie lengths were ready to 240 min for each SMRT cell.
Illumina sequencing and contigs obtained by de novo associates
For the SGS sequencing short reads data(Hiseq) in Arabidopsis (SRR2898686, SRR2898687 and SRR2898688) were downloaded. With those the sequencing raw datasets from P.hybrida, we carried out a stringent filtering process of raw sequencing reads before the transcriptome assembly. Both raw reads of Miseq and Hiseq were cleaned by removing adapter sequences, not-coding RNA (such every bit rRNA, tRNA and miRNA), and low-quality sequences (reads with ambiguous bases 'N'). Hiseq clean reads were assembled with Trinity26 using the following parameters: –edge-thr = 0, –menstruation-thr = 0 and the balance default parameters. This meant keeping the edge as much as possible in Butterfly. Miseq make clean reads were overlapped past Wink-1.ii.6 using the following parameters: -M 200 -r 300 -f 500 -s 50.
Total-length corrected transcripts collection in SMART sequencing
Full-length transcripts which contained poly-A tails, 5′ primers and 3′ primers were obtained past using Pacific Biosciences' SMRT analysis software (v2.3.0). Any reads shorter than 200 bp and those with a predicted accuracy lower than 75 were removed. The accuracy of full-length transcripts was generally not as high as SGS short reads. LSC 0.3.19 was used to correct full-length transcripts. The options "I_nonredundant" was prepare to "N" and "I_RemoveBothTails" was set to "Y". The length of pseudo chromosomes was 50,000,000 and the length of sequence gap between long reads was 100. Minimum number of not'N' after compressing was set to 39, and "maximum 'North' immune afterward compressing" was set as 1.
Mapping finding betwixt corrected PacBio reads and contigs and best-hitting longest contigs option
In our hybrid sequencing and map finding method, the common BLAT alignment algorithm was used, the post-obit hypothesis was proposed, and we formulated half-dozen likelihood functions:
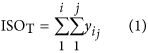
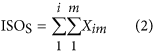
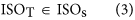
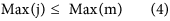
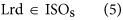
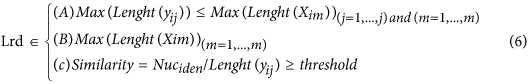
where IsoT is the isoform library divers from the SMRT sequencing, IsoS is the contigs (isoforms) library assembled from short reads based on the Illumina sequencing. The two types of isoforms libraries correspond the isoforms sets of all the expressed genes in the specific cell, tissue or organ, as defined by two different sequencing methods. yij indicates the jth isoform of the ith gene in the isoform library derived from SMRT sequencing; Tenim indicates the mth isoform of the ith gene assembled from curt reads. Part (4) shows that the number of true expressing isoforms of each cistron obtained from SMRT sequencing is less than that of the theoretical associates from curt reads. Lrd represents the set of longest contigs that are selected from the contigs (isoforms) library assembled from short reads (shown in role (five)) and the members of Lrd should meet the demands listed in office (6). Equally shown in function (six), firstly, the length of the mapping isoform of an individual factor in IsoSouth should be equal or longer than those corresponding isoforms derived from SMART sequencing (office A), and the longest one represents the form that contains near all total-length exons of that gene (function B). Secondly, those mapping molecules should have a loftier level of similarity (we advise typically >99% threshold), that is the rate of the identified nucleotides (Nuciden) to the length of the mapped isoform (Length(yij)) in SMRT sequencing (function B). Based on the theoretical hypothesis, we developed a script for performing the map finding process to identify the longest contigs datasets. In addition, the method developed past u.s. was likewise used to conduct out the back-up removal assay in our PacBio corrected long reads as described hither. Thus, if the length of read A was longer than read B, and in improver read B did non accept overhangs, the similarity was higher than 0.99 and in that location were no gaps betwixt the two reads, so we concluded that read B was a duplication of read A, and so deleted the B reads. By adjustment the duplication-removed and corrected long reads (DRCLR) to contigs assembled by Trinity with a 99% threshold, we assigned the longest contigs identified to exist DRCLR, regardless of any gaps in the reference contigs.
Alternative splicing assay
Based on our mapping strategy, after BLAT alignment software was used to marshal DRCLR to the contigs assembled by short reads from SGS (Illumina Hullo-Seq) by Trinity using default parameters, the longest contigs, representing 99% of our defined similarity, were selected to make up the reference library. We divers the alignment gaps longer than fifty bp as splices. Dissimilar lengths or sites of the gaps were divers as alternative splicing.
Isoform differential expression analysis
Isoform expression levels among various samples were identified based on the brusque reads datasets and using the isoforms libraries, yielded from the SMRT sequencing assay and from the contigs associates followed by clustering every bit per traditional RNA-Seq, equally reference sequences. Extracts of four different organs (root, stem, leaf and flower) were used as examples of the analysis in this study. The expression analysis from Illumina reads of dissimilar tissues was carried out with bowtie (v1.1.ane) and rsem (v1.2.9)27 using default parameters. P value < 0.05, FDR < 0.01, and a fold alter equal or greater than 2-fold were used every bit the screening cutoffs for determining extremely significant differential factor expression between two samples. Highly expressed isoforms with root-specific characteristics were used every bit examples to test the effectiveness of the ii parallel abundance estimates for isoform analysis.
Additional Information
How to cite this commodity: Ning, G. et al. Hybrid sequencing and map finding (HySeMaFi): optional strategies for extensively deciphering gene splicing and expression in organisms without reference genome. Sci. Rep. 7, 43793; doi: 10.1038/srep43793 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
-
Mortazavi, A. et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008).
-
Wang, Z., Gerstein, Yard. & Snyder, M. RNA-seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009).
-
Quail, M. A. et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13, 341 (2012).
-
Sharon, D. et al. A single-molecule long-read survey of the homo transcriptome. Nat. Biotechnol. 31, 1009–1014 (2013).
-
Tilgner, H. et al. Accurate identification and analysis of human being mRNA isoforms using deep long read sequencing. G3 (Bethesda) 3, 387–397 (2013).
-
Steijger, T. et al. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 10, 1177–1184 (2013).
-
Eid, J. et al. Real-time Deoxyribonucleic acid sequencing from single polymerase molecules. Science 323, 133–138 (2009).
-
Koren, Due south. et al. Hybrid fault correction and de novo associates of single-molecule sequencing reads. Nat. Biotechnol. thirty, 693–700 (2012).
-
Au, Yard. F. et al. Improving PacBio long read accuracy by short read alignment. PLoS 1 7, e46679 (2012).
-
Gnerre, S. et al. High-quality typhoon assemblies of mammalian genomes from massively parallel sequence information. Proc Natl Acad Sci The states 25(108), 1513–1518 (2011).
-
Chaisson, Grand. J. P. et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517, 608–611 (2015).
-
Li, Q. et al. High-accuracy de novo assembly and SNP detection of chloroplast genomes using a SMRT circular consensus sequencing strategy. New Phytol. 204, 1041–1049 (2014).
-
Barbara, T. et al. Cartography of neurexin alternative splicing mapped by unmarried-molecule long-read mRNA sequencing. Proc Natl Acad Sci USA 111, 1291–1299 (2014).
-
Reddy, A. Culling splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol. 58, 267–294 (2007).
-
Barbazuk, W. B., Fu, Y. & Mcginnis, Grand. M. Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 18, 1381–1392 (2008).
-
Filichkin, S. A. et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20, 45–58 (2010).
-
Zhang, Chiliad. et al. Deep RNA sequencing at unmarried base of operations-pair resolution reveals loftier complexity of the rice transcriptome. Genome Res. 20, 646–654 (2010).
-
Trapnell, C. Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
-
Au, K. F. et al. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res. 38, 4570–4578 (2010).
-
Wang, K. et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 38, e178 (2010).
-
Au, Yard. F. et al. Characterization of the human ESC transcriptome by hybrid sequencing. Proceedings of the National Academy of Sciences 110 (fifty), E4821–E4830 (2013).
-
Zhou, R., Noushin, M. & Adams, K. L. Extensive changes to alternative splicing patterns following allopolyploidy in natural and resynthesized polyploids. Proc Natl Acad Sci United states 108, 16122–16127 (2011).
-
Xu, Z. C. et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Establish J. 82, 951–961 (2015).
-
Chen, L. et al. Transcriptional diversity during lineage commitment of human blood progenitors. Science 345, 1251033–1251033 (2014).
-
Jiang, H. & Wong, W. H. Statistical inferences for isoform expression in RNA-Seq. Bioinformatics 25, 1026–1032 (2009).
-
Grabherr, One thousand. G. et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 29, 644–652 (2011).
-
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq information with or without a reference genome. BMC Bioinformatics 12, e323 (2011).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31572160) and the Primal Inquiry Funds for the Central Universities (2662015PY112). We thank all of the colleagues in our laboratories for constructive word and technical support. Nosotros are as well grateful to Kin Fai Au for providing comments and Dr. Alex C. McCormac for help with editing to the manuscript.
Author data
Affiliations
Contributions
M.N. and 10.C. designed the experiment, analyzed the data and wrote the paper. P.50., F.L. and Z.Westward. performed the experiment and analyzed the data. K.Y., X.L. and D.P. provided assistance in the experiment. G.B. reviewed the manuscript and supervised the whole project. All the authors contributed to discussion and revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution iv.0 International License. The images or other third party material in this article are included in the commodity'due south Creative Commons license, unless indicated otherwise in the credit line; if the cloth is not included under the Creative Commons license, users will demand to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Reprints and Permissions
Most this commodity
Cite this commodity
Ning, K., Cheng, Ten., Luo, P. et al. Hybrid sequencing and map finding (HySeMaFi): optional strategies for extensively deciphering factor splicing and expression in organisms without reference genome. Sci Rep 7, 43793 (2017). https://doi.org/10.1038/srep43793
-
Received:
-
Accepted:
-
Published:
-
DOI : https://doi.org/10.1038/srep43793
Further reading
Comments
By submitting a comment you concord to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag information technology as inappropriate.
Source: https://www.nature.com/articles/srep43793
Belum ada Komentar untuk "Hybrid Sequencing and Map Finding (Hysemafi) Optional Strategies Review"
Posting Komentar